Mark Pickering
Research Projects
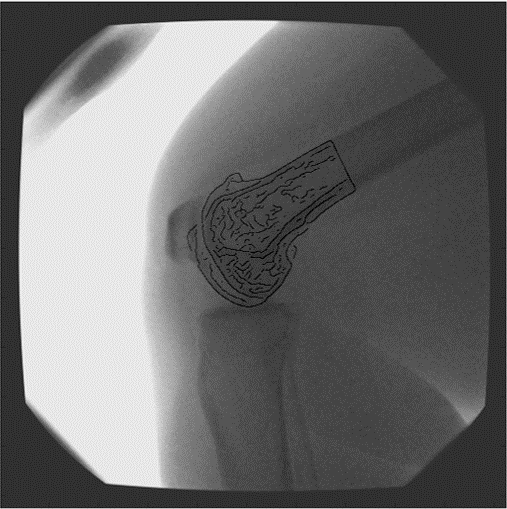
The OrthoVis software is now an intrinsic part of a major clinical trial to evaluate the performance of three different artificial knee designs. The software package has been used to measure the knee motion data for over 170 patients and, when completed, the data collected from the trial will form the largest ever study of knee motion.
The following publications describe methods and experimental results that were generated using the OrthoVis software:
- Saadat S., Perriman D., Scarvell J. M., Smith P. N., Galvin C. R., Lynch J. & Pickering M. R. (2022), An efficient hybrid method for 3D to 2D medical image registration, International Journal of Computer Assisted Radiology and Surgery, Vol. 17, pp. 1313 – 1320, doi: 10.1007/s11548-022-02624-0.
- Saadat S., Asikuzzaman M., Pickering M. R., Perriman D. M., Scarvell J. M. & Smith P. N. (2021) A Fast and Robust Framework for 3D/2D Model to Multi-Frame Fluoroscopy Registration, IEEE Access, Vol. 9, pp. 134223 – 134239, doi: 10.1109/ACCESS.2021.3114366.
- Lynch J. T., Perriman D.M., Scarvell J.M., Pickering M. R., Galvin C. R., Neeman T. & Smith P. N. (2021) The influence of total knee arthroplasty design on kneeling kinematics: A prospective randomized clinical trial, Bone and Joint Journal, Vol. 103, pp. 105 – 112, 10.1302/0301-620X.103B1.BJJ-2020-0958.R1.
- Ward T. R., Hussain M. M., Pickering M. R., Perriman D., Burns A., Scarvell J. & Smith P. N. (2021) Validation of a method to measure three-dimensional hip joint kinematics in subjects with femoroacetabular impingement, HIP International, Vol. 31, pp. 133 – 139, doi: 10.1177/1120700019883548.
- Lynch J. T., Perriman D. M., Scarvell J. M., Pickering M. R., Warmenhoven J., Galvin C. R., Neeman T., Besier T. F. & Smith P. N. (2020) Shape is only a weak predictor of deep knee flexion kinematics in healthy and osteoarthritic knees, Journal of Orthopaedic Research, Vol. 38, pp. 2250 – 2261, doi: 10.1002/jor.24622.
- Lynch, J. T., Scarvell, J. M., Pickering, M. R., Warmenhoven, J., Galvin, C. R., Neeman, T., . . . Perriman, D. (2020). Shape is only a weak predictor of deep knee flexion kinematics in healthy and osteoarthritic knees. Journal of Orthopaedic Research, accepted 30 January.
- Galvin, C. R., Perriman, D., Scarvell, J. M., Lynch, J. T., Pickering, M. R., Smith, P. N., & Newman, P. (2019). Age has a minimal effect on knee kinematics: a cross-sectional 3D/2D image-registration study of kneeling. The Knee, accepted 20 July.
- Ward, T. R., Hussain, M. M., Pickering, M. R., Perriman, D., Burns, A., Scarvell, J., & Smith, P. N. (2019). Validation of a method to measure three-dimensional hip joint kinematics in subjects with femoroacetabular impingement. HIP International. doi:10.1177/1120700019883548
- Scarvell, J. M., Hribar, N., Galvin, C. R., Pickering, M. R., Perriman, D. M., Lynch, J. T., & Smith, P. N. (2019). Analysis of kneeling by medical imaging shows the femur moves back to the posterior rim of the tibial plateau, prompting review of the concave-convex rule. Physical Therapy, 99(3), 311-318. doi:https://doi.org/10.1093/ptj/pzy144
- Akter, M., Lambert, A. J., Pickering, M. R., Scarvell, J. M., & Smith, P. N. (2014). Robust initialisation for single-plane 3D CT to 2D fluoroscopy image registration. Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization, Taylor & Francis, DOI: 10.1080/21681163.21682014.21897649. doi:10.1080/21681163.2014.897649
- Scarvell, J. M., Pickering, M. R., & Smith, P. N. (2009). New registration algorithm for determining 3D knee kinematics using CT and single-plane fluoroscopy with improved out-of-plane translation accuracy. Journal of Orthopaedic Research, 28(3), 334-340.
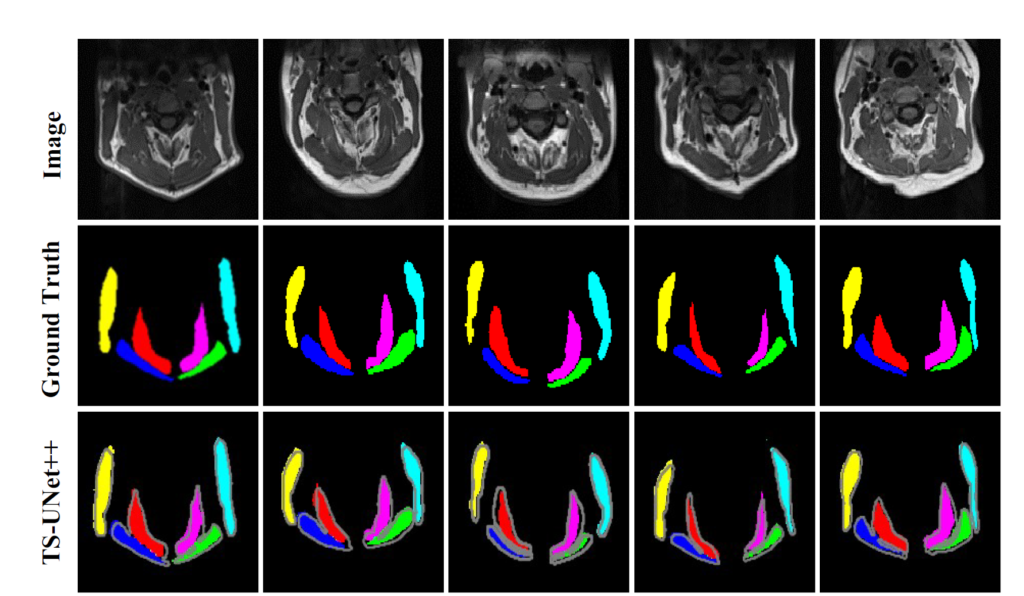
The following publication describes the algorithm and experimental results in more detail:
- Suman AA; Khemchandani Y; Asikuzzaman M; Webb AL; Perriman DM; Tahtali M; Pickering MR, 2020, ‘Evaluation of U-Net CNN Approaches for Human Neck MRI Segmentation’, in 2020 Digital Image Computing: Techniques and Applications, DICTA 2020, doi: 10.1109/DICTA51227.2020.9363385.
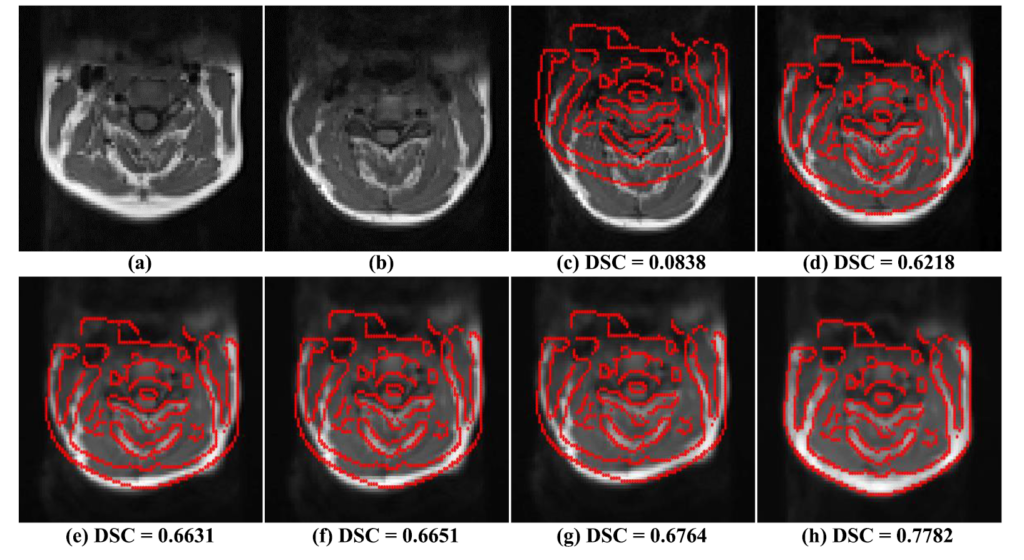
The following publications describe the algorithm and experimental results in more detail:
- Al Suman, A., Asikuzzaman, M., Webb, A. L., Perriman, D. M., Tahtali, M. & Pickering, M. R. (2020). A Deformable 3D-3D Registration Framework Using Discrete Periodic Spline Wavelet and Edge Position Difference. IEEE Access, 8, 146116-146133. doi: 10.1109/ACCESS.2020.3015504.
- Suman, A. A., Aktar, M. N., Asikuzzaman, M., Webb, A. L., Perriman, D. M., & Pickering, M. R. (2019). Segmentation and reconstruction of cervical muscles using knowledge-based grouping adaptation and new step-wise registration with discrete cosines. Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization, 7(1), 12-25. doi:10.1080/21681163.2017.1356751

The following publications describe the algorithm and experimental results in more detail:
- Chandio A. A., Asikuzzaman M., Pickering M. R. & Leghari M. (2022) Cursive Text Recognition in Natural Scene Images Using Deep Convolutional Recurrent Neural Network, IEEE Access, Vol. 10, pp. 10062 – 10078, 10.1109/ACCESS.2022.3144844.
- Chandio, A. A., Asikuzzaman, M., Pickering, M. R. & Leghari, M. (2020). Cursive-Text: A Comprehensive Dataset for End-to-End Urdu Text Recognition in Natural Scene Images. Data in Brief, 31, 105749. doi: 10.1016/j.dib.2020.105749
- Chandio, A. A., Asikuzzaman, M. & Pickering, M. R. (2020). Cursive Character Recognition in Natural Scene Images Using a Multilevel Convolutional Neural Network Fusion. IEEE Access, 8, 109054-109070.